MasSpec Pen for cancer detection
Published on: 26 Sep 2017
Mass spectrometry tissue sampling pen offers surgeons near real-time molecular confirmation of tumor excision
Fully removing tumor tissue is vital in maximizing post-operative disease-free survival for cancer patients. At present, it can be difficult for surgeons to tell whether they have fully removed cancerous tissues. That’s why researchers at the University of Texas have developed a new handheld tissue sampling device for mass spectrometers that could facilitate surgeons in achieving cancer free “negative margins” when excising tumors. The work has been published in Science Translational Medicine1.
The MasSpec Pen collects molecular information by placing a single water droplet in contact with the tissue surface for just a few seconds (Figure 1). Potential cancer biomarkers including metabolites, lipids and proteins diffuse into the droplet which is automatically withdrawn from the surface and sent directly to a mass spectrometer, for analysis.
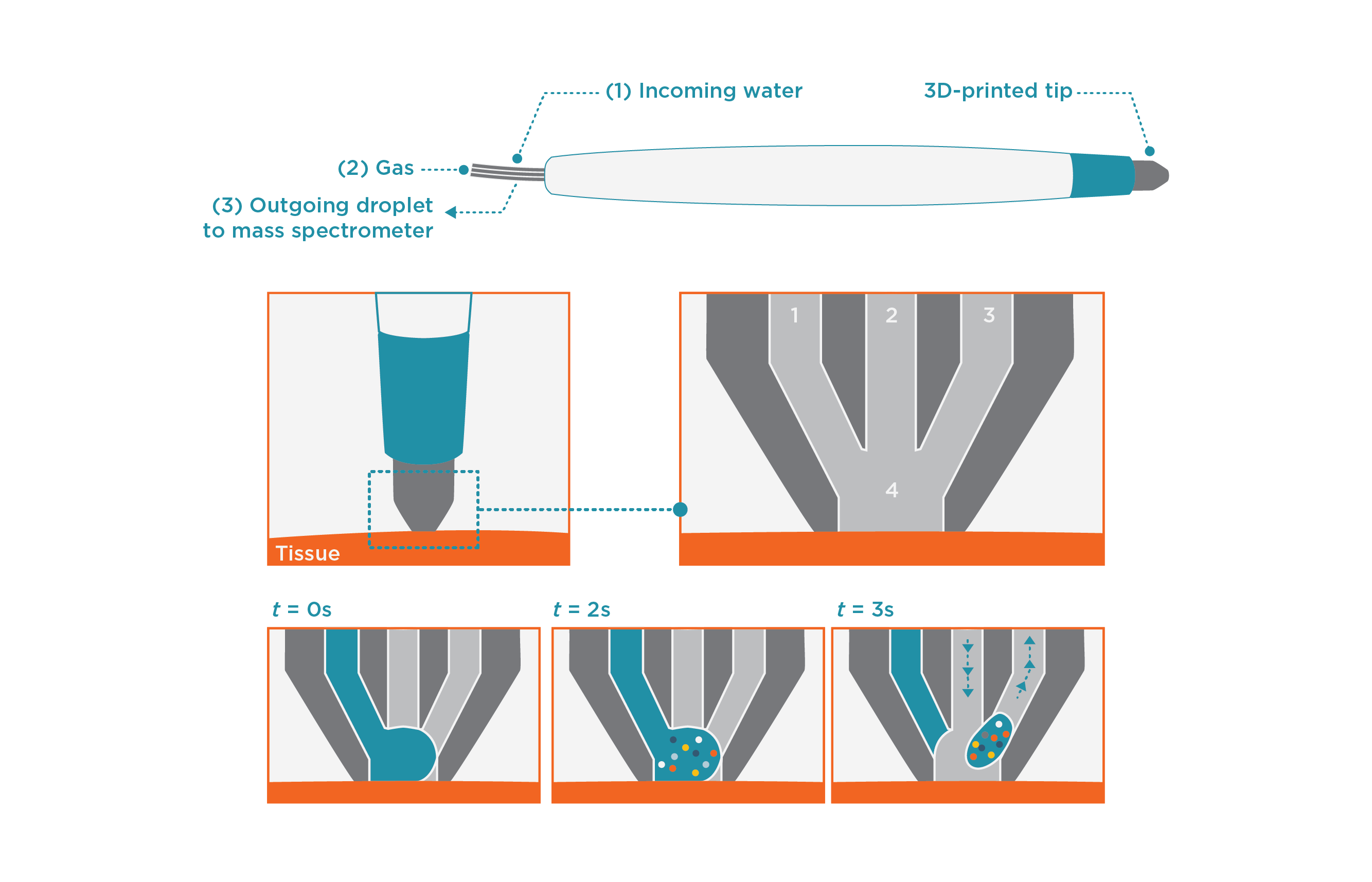
Figure 1. The handheld MasSpec pen is operated using a foot pedal, which activates an automated sequence of droplet exposure, transport and analysis by mass spectrometer. The system also automatically cleans itself by flushing between samplings.
Applying only water, the Pen causes no measurable tissue damage. This makes it ideal for using in vivo, which the team demonstrated by analyzing the tissue of living mice without causing damage or measurable stress to the animal.
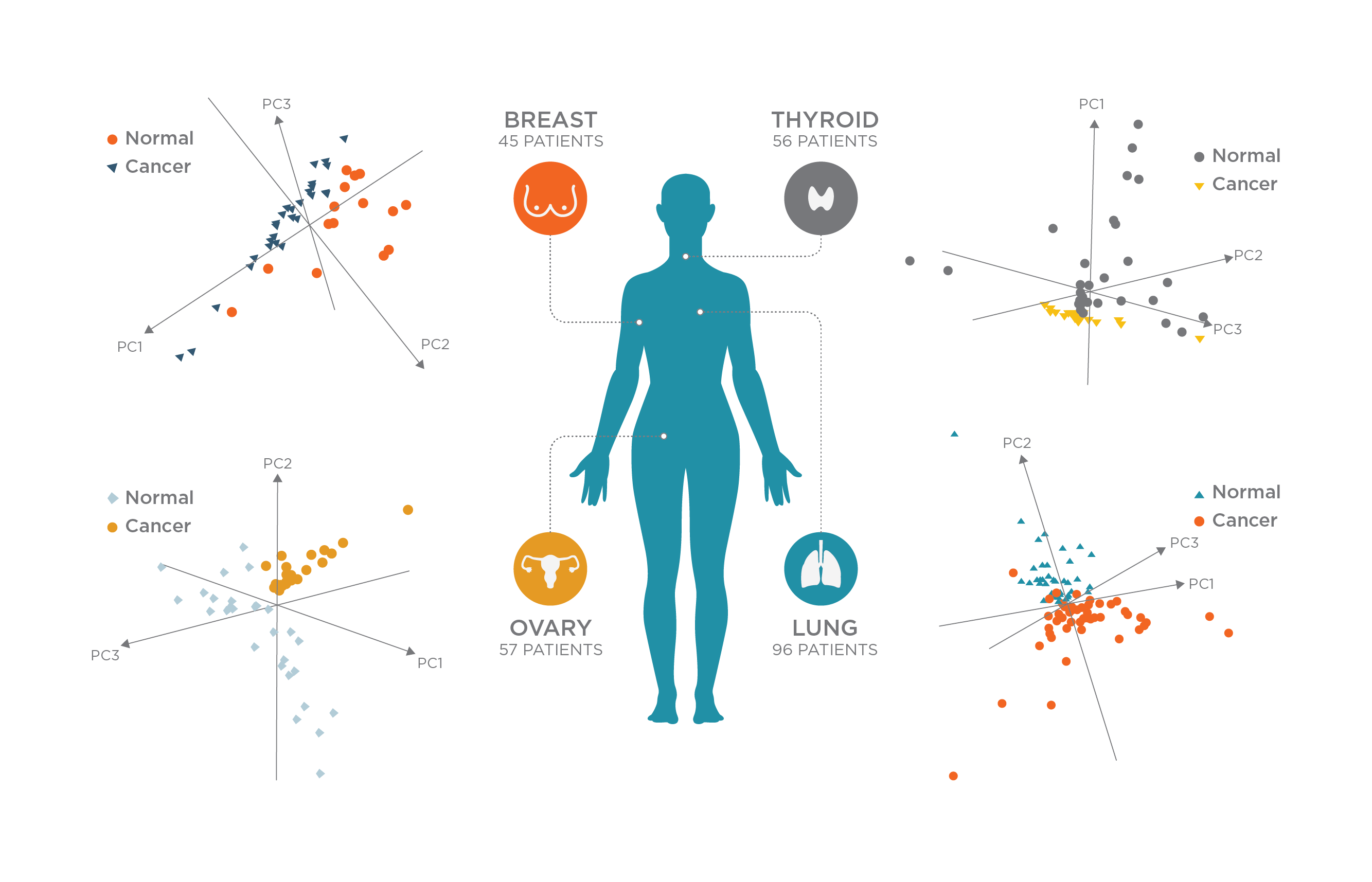
To test whether the Pen system was able to accurately identify tumor tissues, the authors analyzed hundreds of healthy and cancerous ex vivo human tissue samples (Figure 2). Statistical classifiers built using machine learning algorithms and histologically validated molecular information were used to detect cancer using data collected for the sampled tissues. The classifiers were able to detect cancer successfully with an overall sensitivity of 96.4%, specificity of 96.2%, and accuracy of 96.3%.
The classifiers were also able to detect cancer in marginal tumor regions presenting mixed histologic composition, the areas that present most difficulty for surgeons.
Using mass spectrometry for the analysis of tissues offers benefits over other near real-time solutions for in vivo cancer detection. Many of the other technologies in this space rely on injection of exogenous chemical labels that target specific cell types, allowing visualization of tumor margins. They also often require excision prior to ex vivo processing and analysis, and can suffer from poor tissue specificity.
Mass spectrometry returns much more biochemical information, allowing observation of panels of chemical biomarkers for use in powerful statistical analyses. A key challenge for this technology is developing smaller, cost-effective mass spectrometers with enough analytical performance to identify the biomarkers in question.
References
- Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system, J. Zhang, J. Rector, J.Q. Lin, J.H. Young, M. Sans, N. Katta, N. Giese, W. Yu, C. Nagi, J. Suliburk, J. Liu, A. Bensussan, R.J. DeHoog, K.Y. Garza, B. Ludolph, A.G. Sorace, A. Syed, A. Zahedivash, T.E. Milner and L.S. Eberlin, Science Translational Medicine, 9, 406, 2017, DOI: 10.1126/scitranslmed.aan3968
Interview – Prof. Livia S. Eberlin, University of Texas at Austin
You recently published a spectacular paper in Science Translational Medicine. Can you summarize the most important findings of this research?
The most important findings are that we developed an automated, handheld and biocompatible device that can be used to diagnose cancer based on molecular information without causing any measurable harm to the tissue sample. We demonstrated the performance ex vivo and in vivo and believe our results support the idea that this system could be incorporated in clinical practice in a variety of surgical modalities without significantly interfering with surgical workflow.
Using the mass spectra you were able to identify several potential cancer biomarkers including metabolites, lipids and proteins. Do your classifiers incorporate information from all types of molecules? Or which are the most useful as biomarkers in your opinion?
Lipids have been the most useful biomarkers acquired by ambient ionization mass spectrometry for cancer diagnosis. Our statistical classifier identified a sub-set of the detected molecules as predictive markers, and the majority of them are lipids.
Did you identify different biomarkers for the different tumor types you looked at, or was there some overlap between tumor types?
The molecules are often similar, but the relative intensities or the mass spectra profiles are different for each tumor type.
Can the statistical analysis of cancer biomarkers be done in real-time, or is the data analysis a time limiting step?
That is certainly not the time-limiting step. We’ve developed software that can provide statistical analysis in under a second.
What will be the next steps needed before surgeons can use this technology in the clinic?
We are starting to test the approach during surgeries in clinic early next year. It will take a while to go through all the regulatory steps to turn this into a medical device.
Do you have in mind any smaller, more portable analytical instruments that could potentially offer an alternative to mass spectrometry, facilitating application of the MasSpec pen at point-of-care?
Absolutely, that is something we want and are currently exploring.
Can you briefly tell us about your future plans for this research?
We plan to expand to other cancer types and further validate our results. Then we will start testing in surgeries to properly evaluate its performance in surgical margin evaluation.
