Gut Microbiome Phenotyping Using Dynamic Breath Analysis
Published on: 17 Nov 2023
Metabolomic studies involve probing the metabolites produced by the physiological processes in our body such as cellular metabolism to investigate health and disease. Many metabolites that are produced in our body can have an effect on our systems and many are produced by the community of microbes that live in our intestinal tract, known as the gut microbiome. The gut microbiome interacts with the food that we eat by using the nutrients we provide to generate their own energy and undergo their own metabolic processes – which release metabolic byproducts that can have both beneficial and deleterious effects on us.
Disturbances or an unbalancing of the gut microbiome (dysbiosis) can result in the excessive production of harmful metabolites that can exacerbate or contribute to the development of many chronic diseases such as liver disease (1,2), cardiovascular disease (3), inflammatory bowel disease (IBD) (4), colorectal cancer (CRC) (5,6), and many more. Many of these metabolites are volatile, and so can diffuse from their point of origin in the gastrointestinal tract into the bloodstream.
Upon reaching the alveoli in the lungs, volatile metabolites are exchanged into the air and exhaled in the breath. The concentration of breath metabolites is proportional to their blood concentration (7), and so gut microbiome-originating volatile metabolites that are important for health and disease can be measured non-invasively in exhaled breath.
Dr. Giuseppe Ferrandino examines the ability of dynamic breath analysis to non-invasively profile the gut microbiome in his BBCon23 talk ‘Gut Microbiota Profiling via Dynamic Breath Analysis:
If the video fails to load please ensure you have cookies enabled.
Testing the feasibility of this approach in a pilot study:
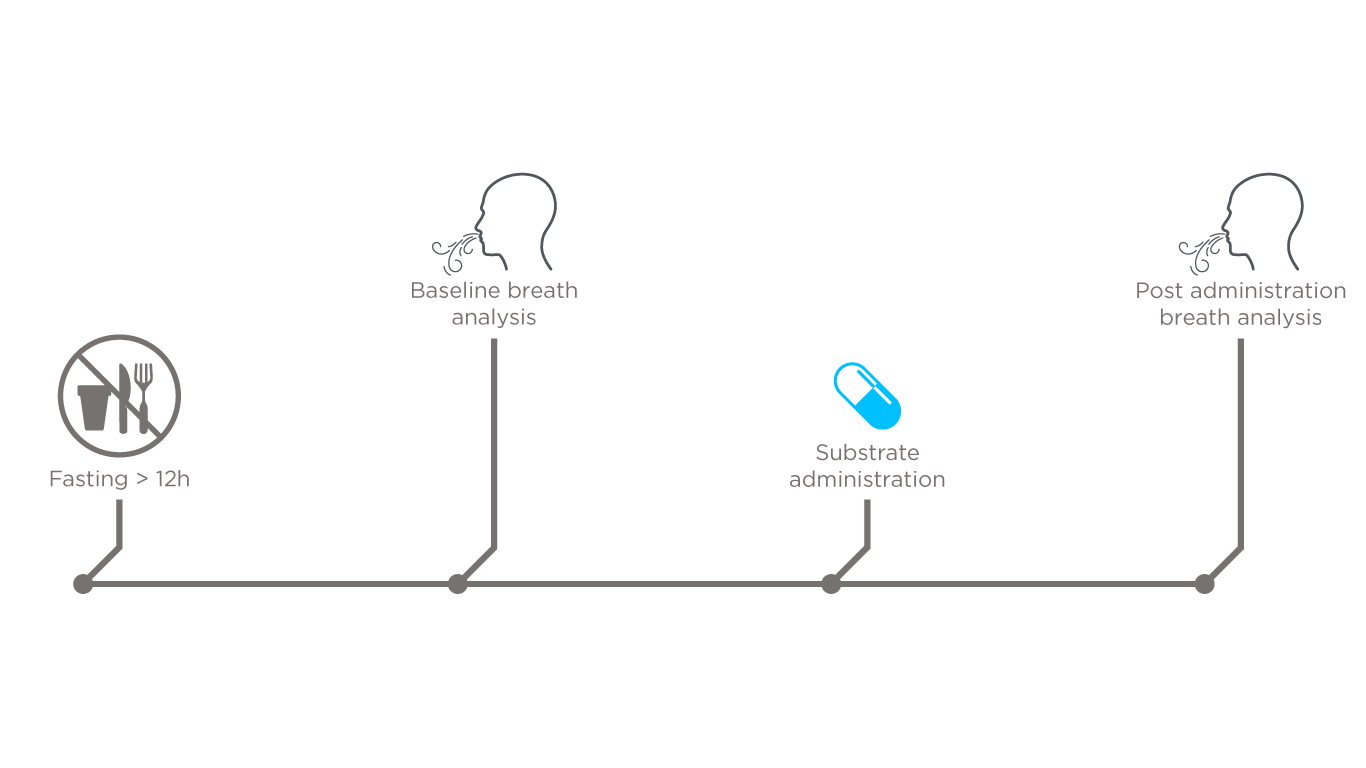
In order to test the feasibility of this approach to study gut microbiome metabolites, we designed a pilot study to analyze breath composition before and after the administration of a test substrate (75g of glucose). A total of 10 healthy volunteers were enrolled for this study and instructed to fast overnight. Next, breath was analyzed directly prior to and post substrate administration using selected ion flow tube mass spectrometry (SIFT-MS). Each subject breathed into the instrument for approximately 15 seconds every 5 minutes up until 90 minutes after the glucose was ingested. This allowed for the detailed mapping of the changing composition of breath over time.
Results:
We measured several metabolites in exhaled breath that were reported to be generated by the gut microbiome (summarized in Table 1).

The major focus of this study was the fermentation products of carbohydrates which are metabolites generated via ethanol and 2-3-butanediol fermentation, given both these pathways have been associated with non-alcoholic steatohepatitis (NASH), and non-alcoholic fatty liver disease (NAFLD). There was a spike of ethanol in the breath in all tested subjects, showing variability either in the duration or in the extent of the spike, suggesting a highly personalized response.
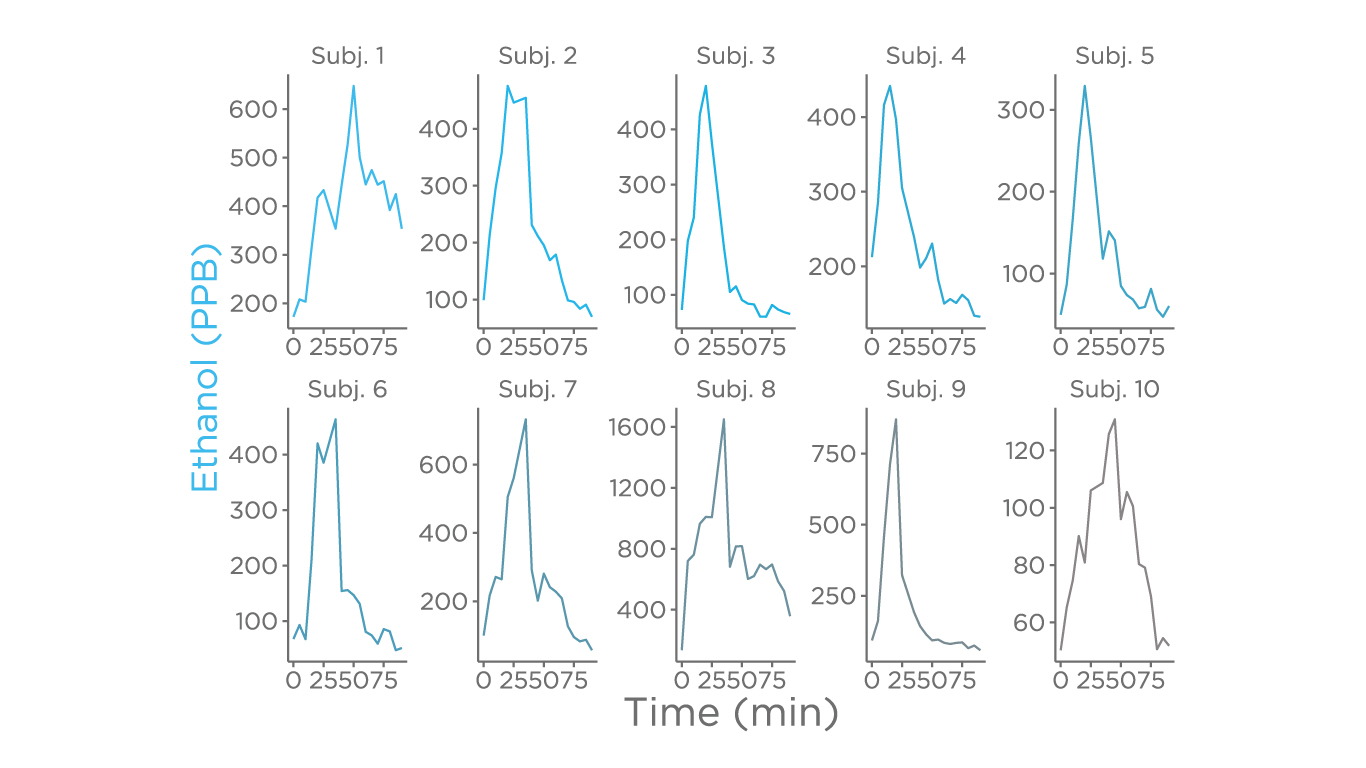
We observed a similar response for acetoin (an intermediate of the 2-3-butanediol fermentation), and propanoic acid (a short-chain fatty acid that is generated by the gut-microbiome metabolism of carbohydrates). Currently, hydrogen and methane in breath are used as biomarkers for gut microbiome bioproducts for the diagnosis of short intestine bacterial overgrowth (SIBO), intestinal methanogen overgrowth (IMO), and carbohydrate malabsorption (CM) (8,9). As we have demonstrated in this study, many other metabolites generated by the gut microbiome and associated with different diseases are also detectable in breath. Future studies will expand our focus to metabolites generated by microbiome amino acid metabolism, where their involvement in chronic diseases is just emerging (10).
Levels of hydrogen and methane are measured in breath after specific substrate administration such as lactulose or glucose for SIBO testing, or fructose and lactose for CM testing. It is possible that by utilizing different test substrates, different microbial metabolic processes could be probed and informative changes in breath composition analyzed. This is known as an exogenous volatile organic compound (EVOC) probe approach which we believe could significantly increase the power of breath analysis.
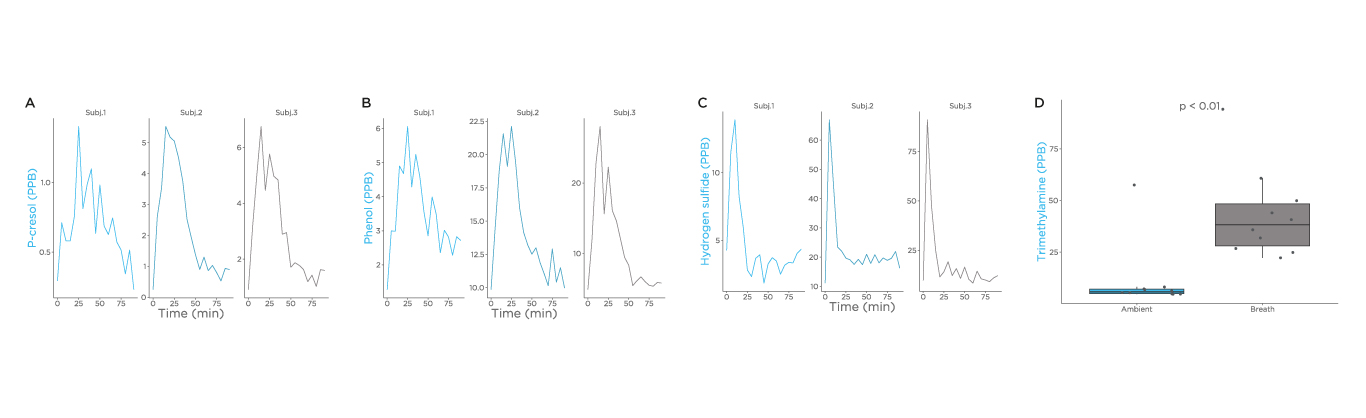
Additional substrates that were investigated as part of this study were choline, tryptophan, and cysteine (Fig), and we observed a spike of cresol, phenol, and hydrogen sulfide. Although trimethylamine can be measured in breath, a spike was not observed after administration within the 90-minute time frame during this study. Different test substrates and a longer time frame for analysis are important future directions for this research.
What are the benefits of using breath for gut microbiome studies?
This non-invasive method for probing gut-microbiome-originating metabolites supports breath as a feasible platform for metabolomic analysis. Like other biological matrices such as urine and blood, breath has its own strengths and limitations for the analysis of compounds for the purpose of biomarker measurement. One of the most immediately apparent is that breath collection can be undertaken in a convenient and completely non-invasive manner. Breath sampling is comfortable, and acceptable to patients or trial participants. A comfortable mode of sample collection has multiple benefits beyond just the improved comfort for patients and trial participants, including easier ethical approval during clinical trials, and higher uptake for screening programs.
As breath is a virtually inexhaustible resource produced almost continually from the body, a large volume of breath can be collected at any one time. The volatile compounds in breath samples can also be pre-concentrated prior to analysis, which can ensure the capture of even very low-abundance chemicals. Theoretically, breath sampling can be undertaken anywhere, enabling decentralized trial designs and at-home breath testing for disease monitoring and diagnosis. Therefore, breath analysis can be used to identify the role of the gut microbiome in health and in disease.
Owlstone Medical can help you incorporate breath analysis into your research
Owlstone Medical is the world leader in breath analysis, and we can help you incorporate breath into your microbiome research. As demonstrated in this pilot study, we can use different test substrates to elicit the production of microbial metabolites from specific pathways of interest in an EVOC probe approach. This can be used to study the activity of the microbiome, for example in response to drug treatments, dietary changes, across different demographics (such as age), or in health and disease states. See the poster of this pilot study for more details or read our microbiome 101 document for a summary of what microbiome-associated compounds we can study in breath and what products and services we can offer.
View the Poster View the Microbiome 101
References:
1. Yuan J, Chen C, Cui J, Lu J, Yan C, Wei X, et al. Fatty Liver Disease Caused by High-Alcohol-Producing Klebsiella pneumoniae. Cell Metab. 2019 Oct 1;30(4):675-688.e7. DOI: 10.1016/j.cmet.2019.08.018
2. Meijnikman AS, Davids M, Herrema H, Aydin O, Tremaroli V, Rios-Morales M, et al. Microbiome-derived ethanol in nonalcoholic fatty liver disease. Nat Med. 2022 Oct;28(10):2100–6. DOI: 10.1038/s41591-022-02016-6
3. Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011 Apr;472(7341):57–63. DOI: 10.1038/nature09922
4. Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020 Apr;17(4):223–37. DOI: 10.1038/s41575-019-0258-z
5. Duizer C, de Zoete MR. The Role of Microbiota-Derived Metabolites in Colorectal Cancer. Int J Mol Sci. 2023 Apr 28;24(9):8024. DOI: 10.3390/ijms24098024
6. Coleman OI, Haller D. Microbe–Mucus Interface in the Pathogenesis of Colorectal Cancer. Cancers (Basel). 2021 Feb 4;13(4):616. DOI: 10.3390/cancers13040616
7. Mochalski P, King J, Mayhew CA, Unterkofler K. Modelling of Breath and Various Blood Volatilomic Profiles-Implications for Breath Volatile Analysis. Molecules. 2022 Apr 7;27(8):2381. DOI: 10.3390/molecules27082381
8. Rezaie A, Buresi M, Lembo A, Lin H, McCallum R, Rao S, et al. Hydrogen and Methane-Based Breath Testing in Gastrointestinal Disorders: The North American Consensus. Am J Gastroenterol. 2017 May;112(5):775–84. DOI: 10.1038/ajg.2017.46
9. Pitcher CK, Farmer AD, Haworth JJ, Treadway S, Hobson AR. Performance and Interpretation of Hydrogen and Methane Breath Testing Impact of North American Consensus Guidelines. Dig Dis Sci. 2022;67(12):5571–9. DOI: 10.1007/s10620-022-07487-8
10. Hoyles L, Fernández-Real JM, Federici M, Serino M, Abbott J, Charpentier J, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med. 2018 Jul;24(7):1070–80. DOI: 10.1038/s41591-018-0061-3
Microbiome Content Pack: exclusive content on how breath analysis can be used for gut microbiome research
